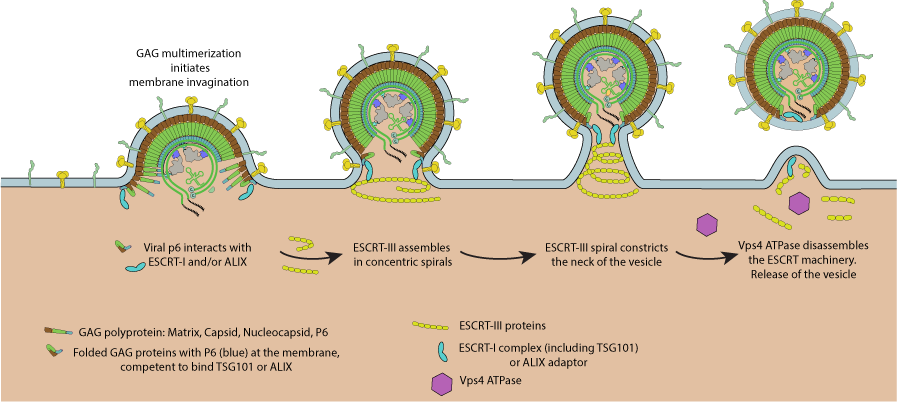
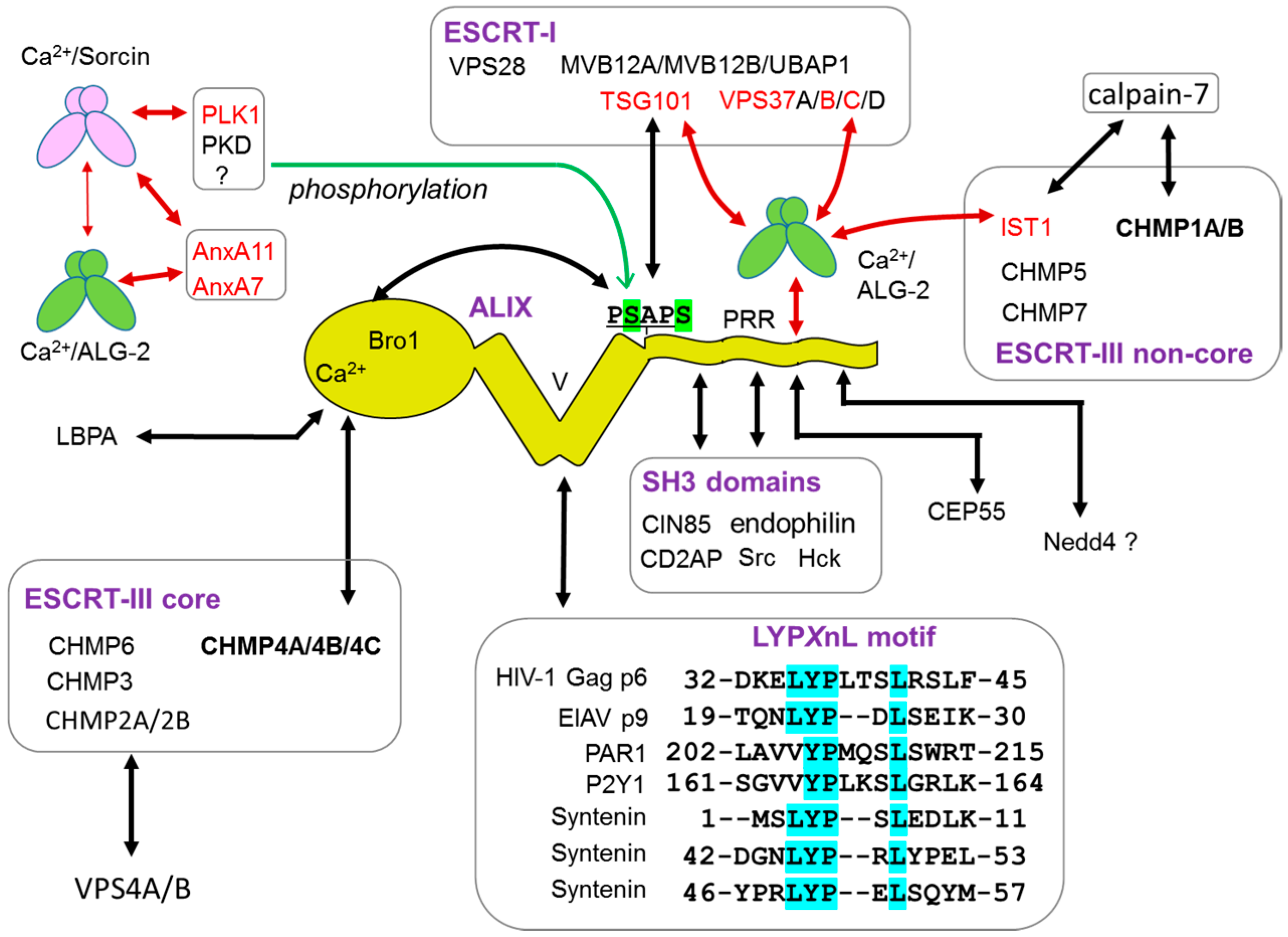
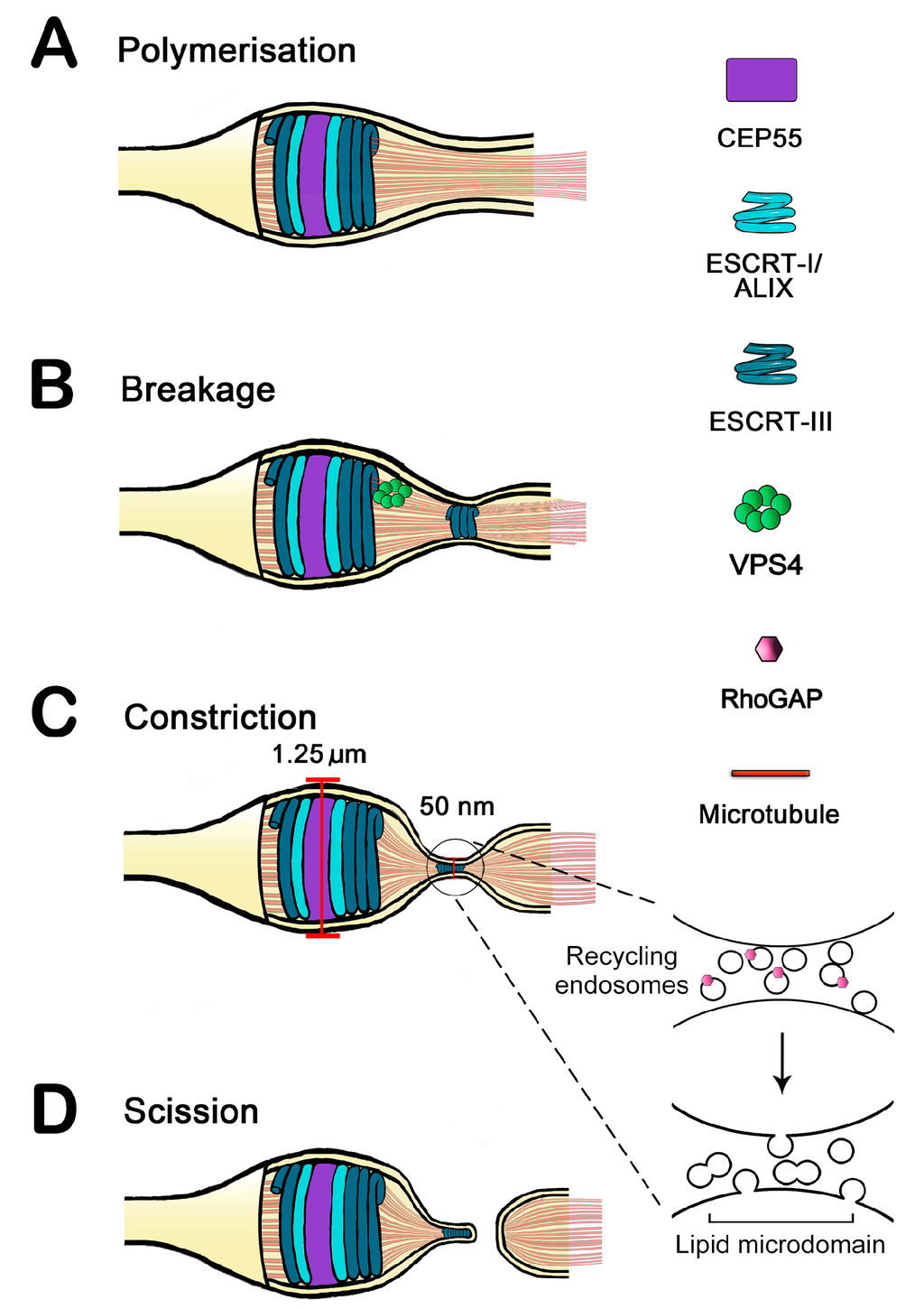
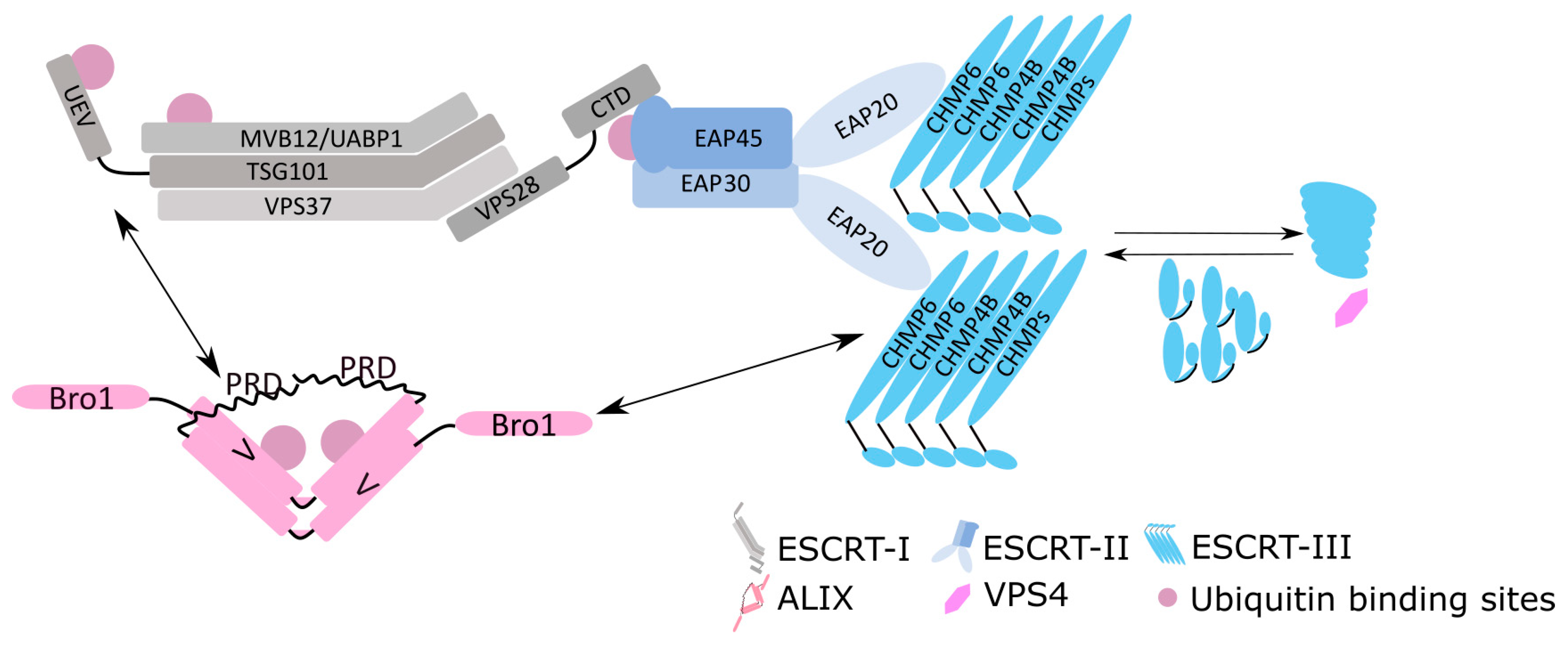
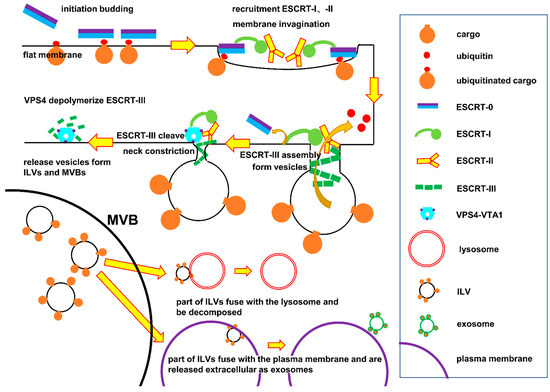
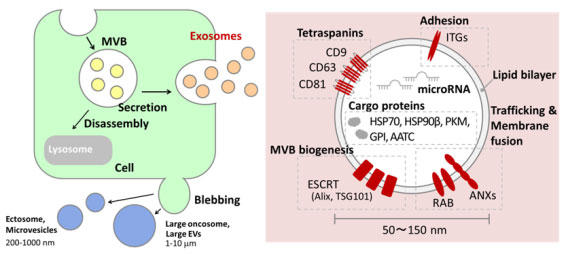
Images of ESCRT
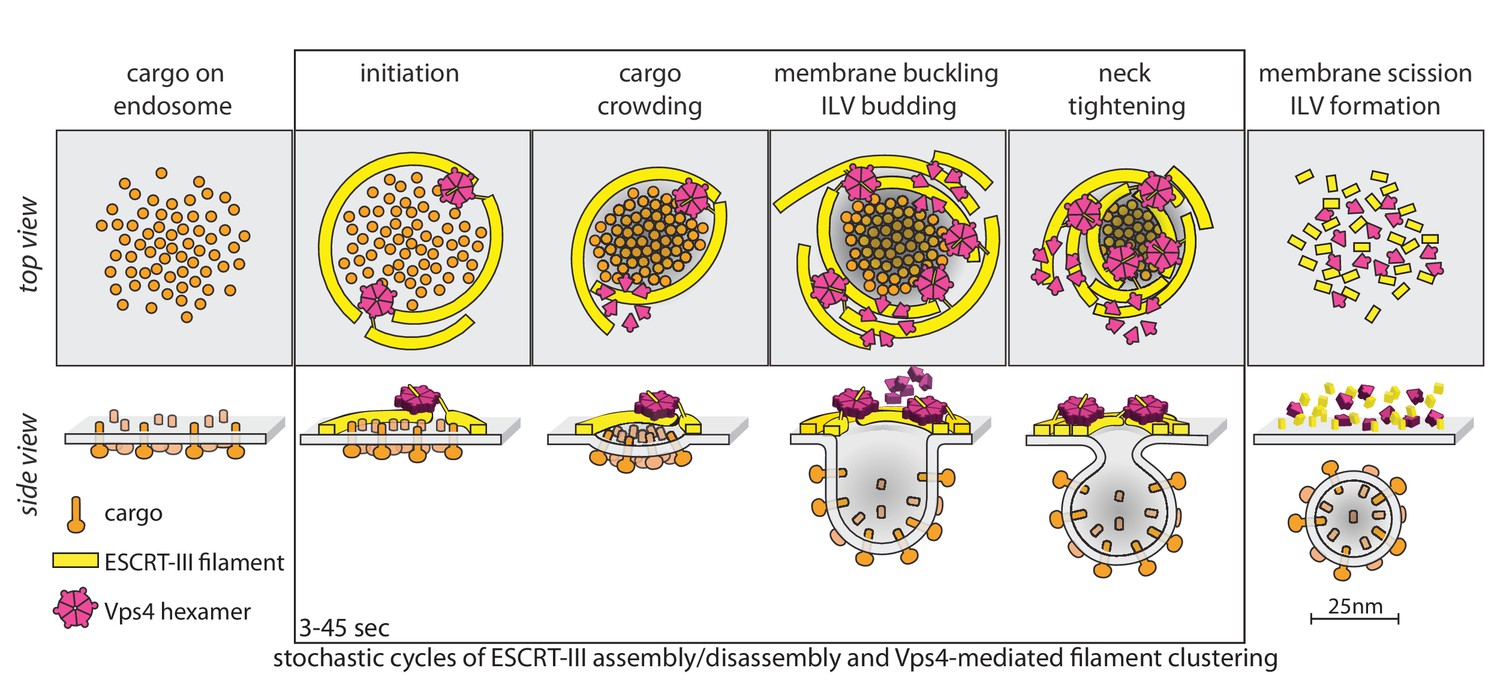
Recruitment dynamics of ESCRT-III and Vps4 to endosomes and implications for reverse membrane budding Formation and three-dimensional architecture of Leishmania adhesion in the sand fly vector ErbB signalling is a potential therapeutic target for vascular lesions with fibrous component Transcriptional regulation of Sis1 promotes fitness but not feedback in the heat shock response
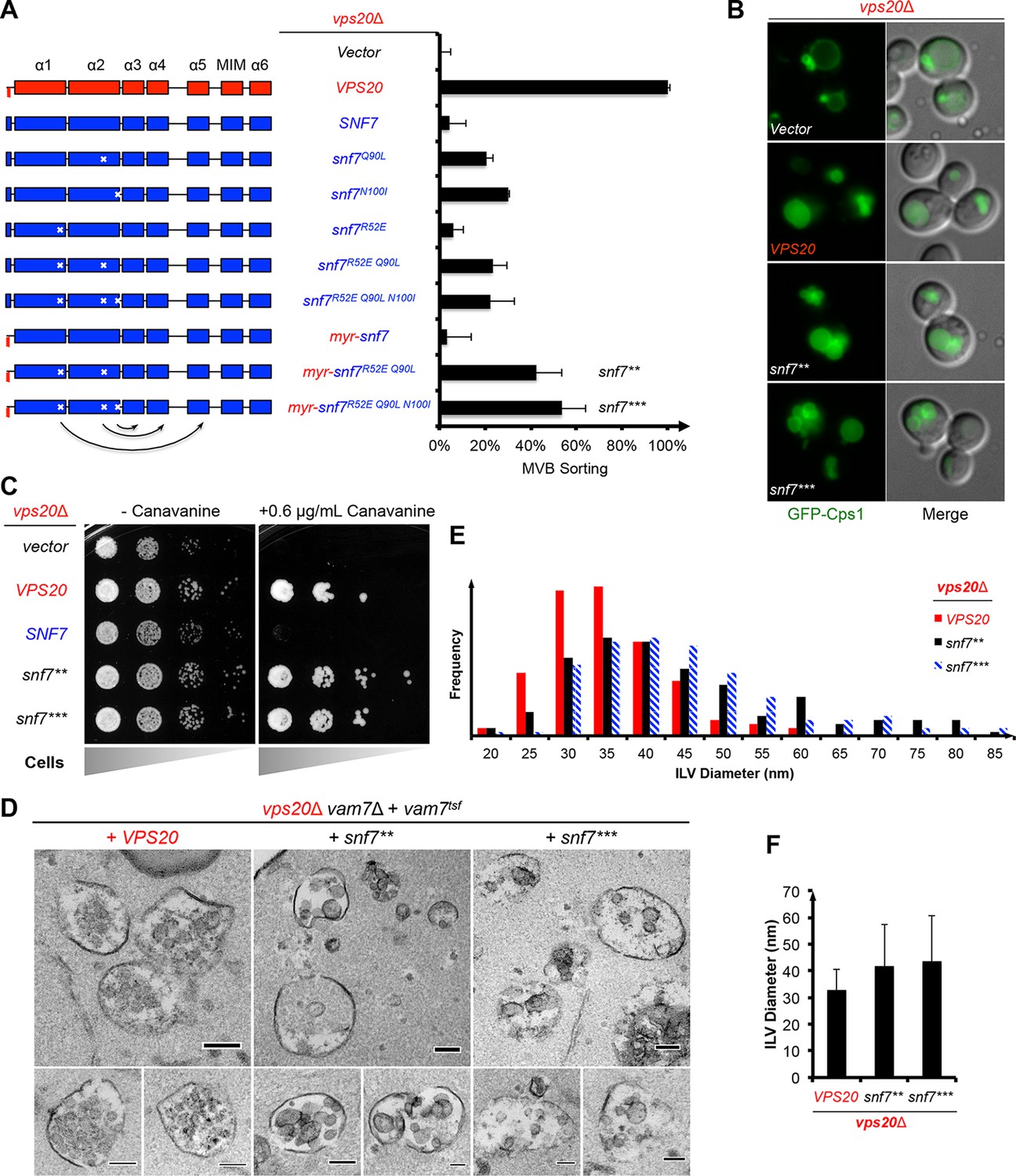
ESCRT-III activation by parallel action of ESCRT-I/II and ESCRT-0/Bro1 during MVB biogenesis Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments Ribozyme activity modulates the physical properties of RNA–peptide coacervates Phase separation-mediated actin bundling by the postsynaptic density condensates
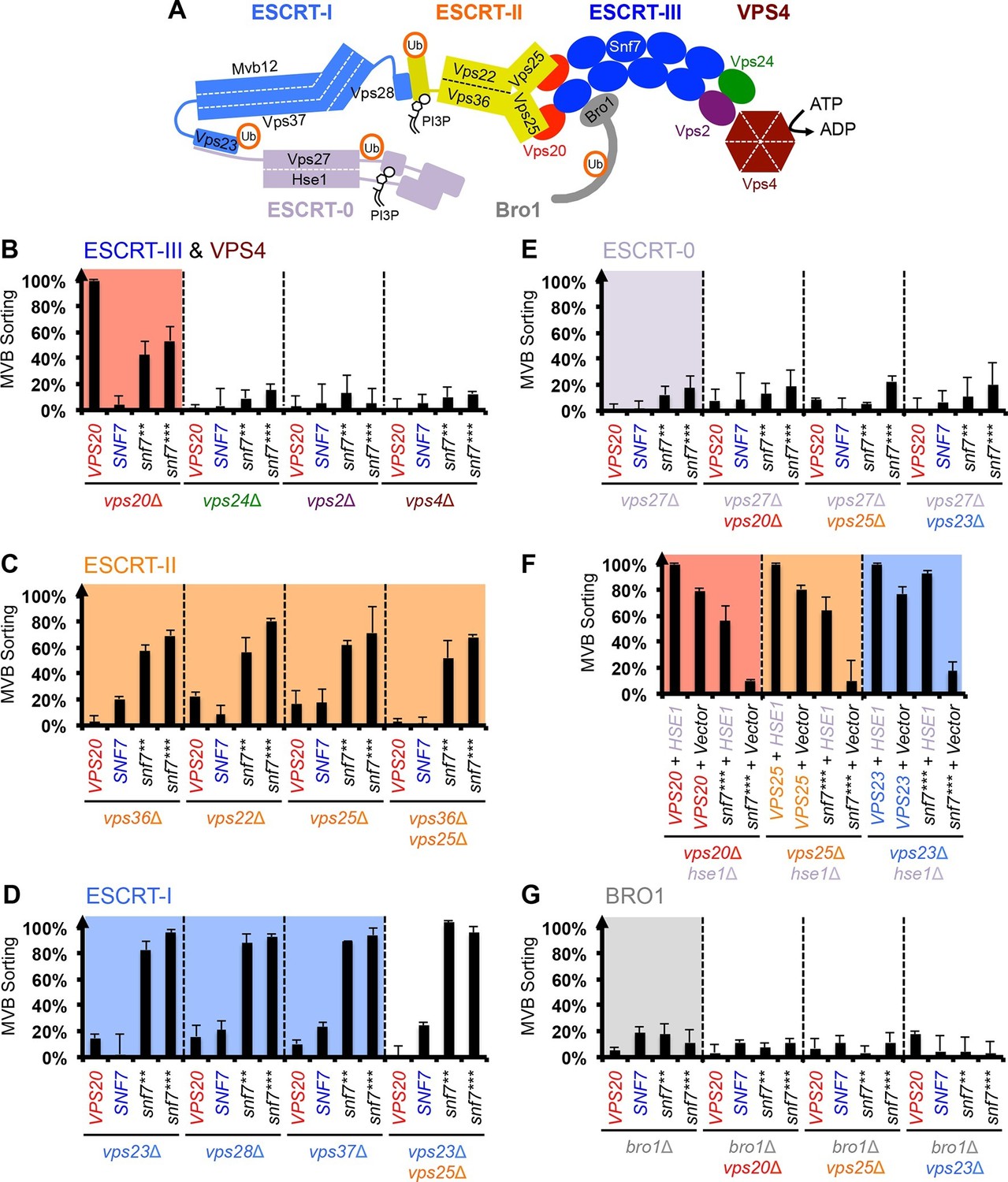
ESCRT-III activation by parallel action of ESCRT-I/II and ESCRT-0/Bro1 during MVB biogenesis Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments Ribozyme activity modulates the physical properties of RNA–peptide coacervates Phase separation-mediated actin bundling by the postsynaptic density condensates
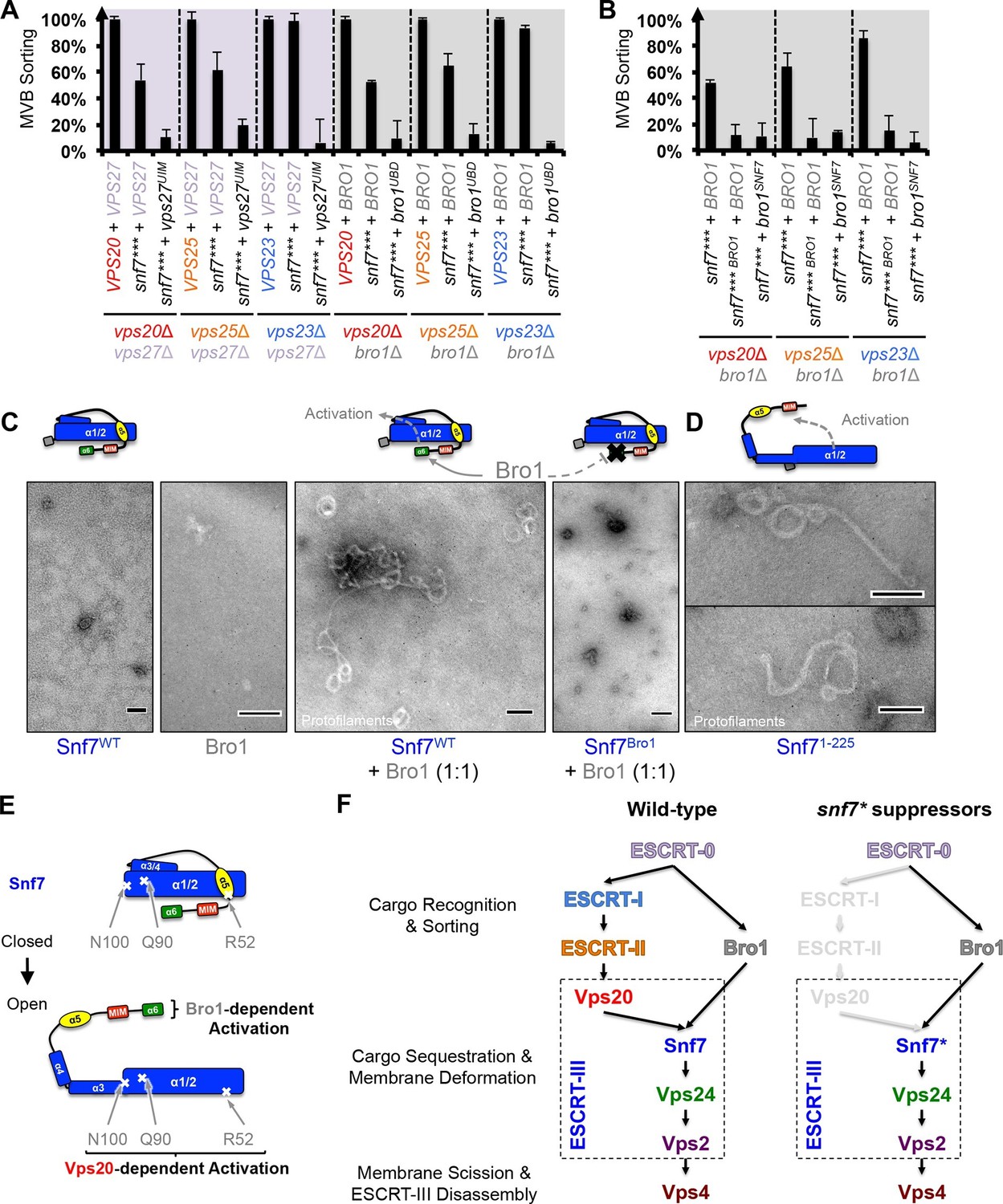
ESCRT-III activation by parallel action of ESCRT-I/II and ESCRT-0/Bro1 during MVB biogenesis Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments SIMMER employs similarity algorithms to accurately identify human gut microbiome species and enzymes capable of known chemical transformations Autophagosome membrane expansion is mediated by the N-terminus and cis-membrane association of human ATG8s
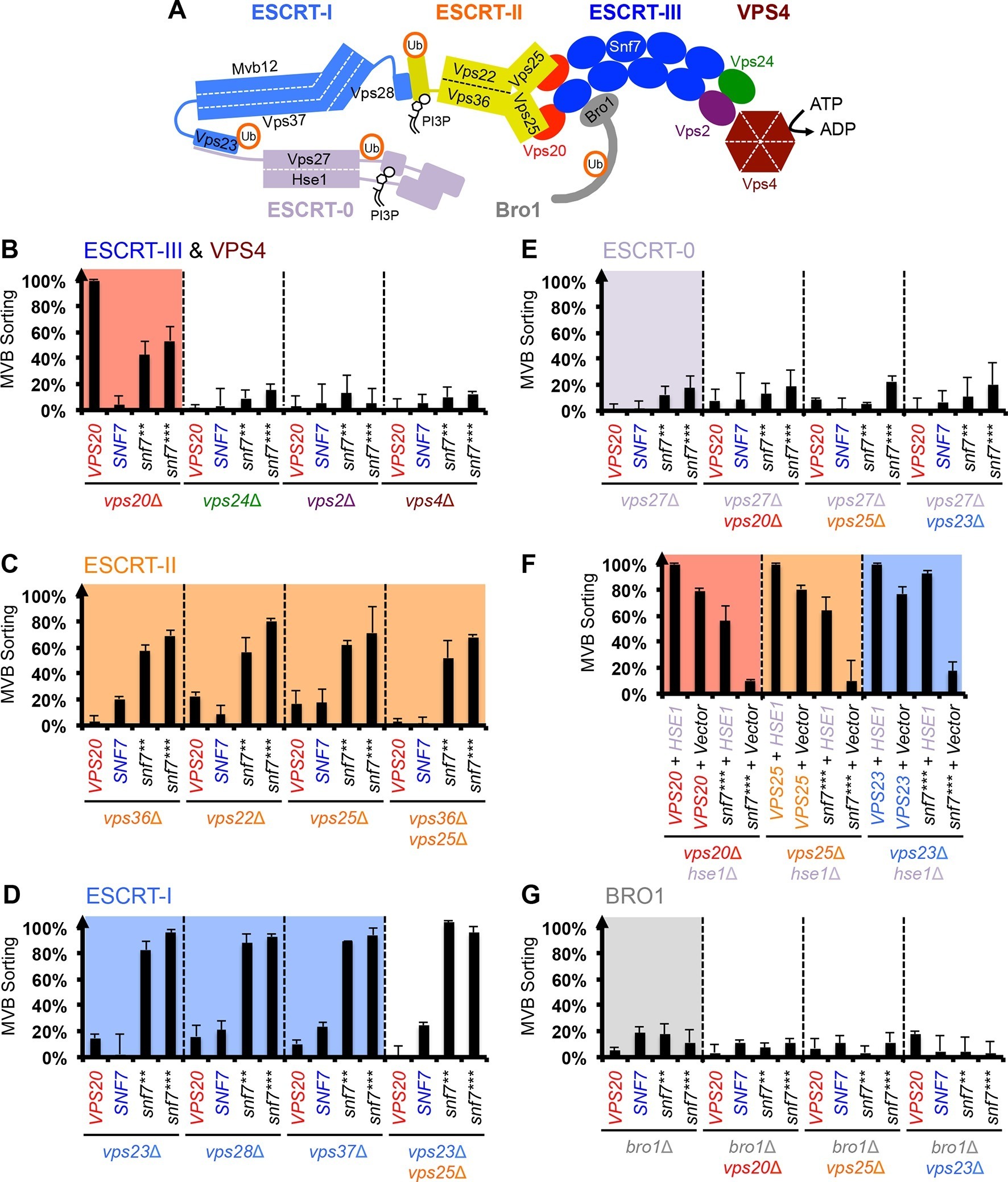
ESCRT-III activation by parallel action of ESCRT-I/II and ESCRT-0/Bro1 during MVB biogenesis Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments SIMMER employs similarity algorithms to accurately identify human gut microbiome species and enzymes capable of known chemical transformations Autophagosome membrane expansion is mediated by the N-terminus and cis-membrane association of human ATG8s
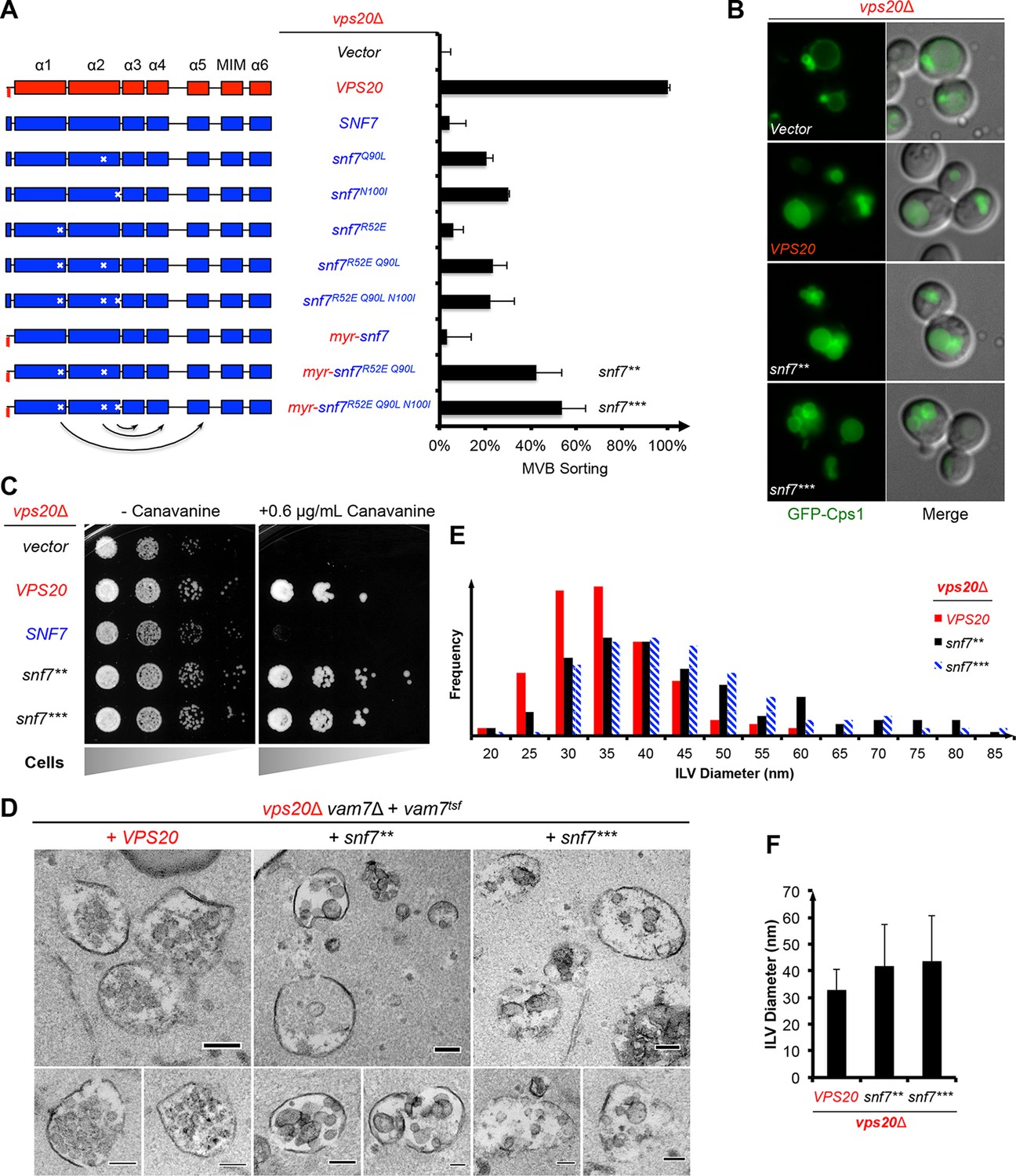
ESCRT-III activation by parallel action of ESCRT-I/II and ESCRT-0/Bro1 during MVB biogenesis Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments Mapping the architecture of the initiating phosphoglycosyl transferase from S. enterica O-antigen biosynthesis in a liponanoparticle A methylation-phosphorylation switch controls EZH2 stability and hematopoiesis

![[新品未開封|SIMフリー] iPhone 16 128GB 256GB 512GB 各色 スマホ 本体](https://thumbnail.image.rakuten.co.jp/@0_mall/icockaden/cabinet/10962912/11261694/iphone16-all.jpg?_ex=300x300)